Current Research
|
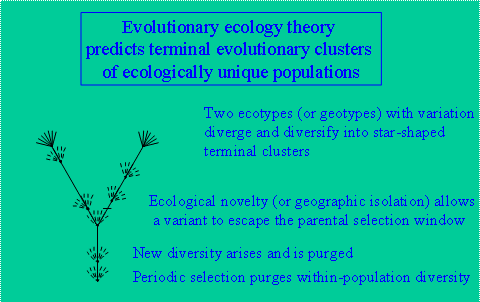
After observing these patterns of diversity and its distribution and their correspondence to ideas about plant and animal speciation, I took a sabbatical leave in 2000 to the lab of Fred Cohan to learn about evolutionary theory. The empirical results we observed in our studies match well with periodic selection theory [Cohan, 2002] (Fig. 9). The idea is that populations, which have variation, evolve through a succession of periodic sweeps of diversity. One most-fit variant out-competing all others and carrying forward the genetic know how to occupy the population’s niche. However, whenever a variant arises which has a novel ecology (occupies a different niche than the parental population) it is no longer selected in the same way (i.e., survives periodic sweeps of the parental population). It is then free to diverge from the parental population, giving rise to a new population which undergoes its own private periodic sweeps. The eventual result is two ecologically distinct populations. Geographic isolation can have the same effect as ecological adaptation in driving populations apart. Fred has shown that a high-resolution molecular technique for analyzing population genetics has potential to detect these terminal ecotypical clusters. The method, called Multi-Locus Sequence Typing (MLST), involves PCR amplification of 7 rapidly evolving protein encoding genes and sorting variants into clonal complexes. Fred has developed an evolutionary simulation that suggests that MLST clonal complexes equate to putative ecotypes. In a project sponsored by the NSF Frontiers in Integrative Biological Research (FIBR) and NASA Exobiology Programs, Fred and I plan to develop a cultivation-independent MLST approach to study, at very high resolution, Synechococcusecotypes within the mat. |
As a part of the FIBR project, we will collaborate with John Heidelberg of the Institute for Genomic Research (TIGR) to obtain genomic sequences of two Synechococcus isolates that are genetically relevant to the mats we study. John will also conduct
direct environmental genomic analysis of predominant mat populations, as a theory-independent
means of investigating how genomic diversity may be organized into populations. Genomic
sequence data will permit us, in collaboration with Devaki Bhayaand Arthur Grossman (Carnegie Institution/Stanford), to develop microarrays that will be used to investigate
gene expression in situ within the mats. Ultimately, we hope to compare the distributions
of allelic variants of highly expressed genes, with alleles that mark MLST clonal
complexes (putative ecotypes).
Figure 9. Periodic Selection Theory.
|