Have we detected all the ecotypes?
We have determined that the 16S rRNA gene may be too conserved to enable us to detect all ecologically distinct populations within microbial communities. At a 68°C site, we also see distinct differently pigmented Synechococcus populations near the mat surface and at depth. However, we can detect no difference in depth in 16S rRNA sequence. We developed an approach to PCR amplify the faster-evolving internal transcribed spacer (ITS) region adjacent to the 16S rRNA gene and used it to show that genotypes found at the mat surface (●) are genetically distinct from those in the mat subsurface (■) (Fig. 6). [Ferris et al., 2003]
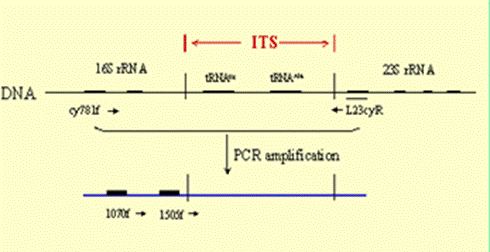
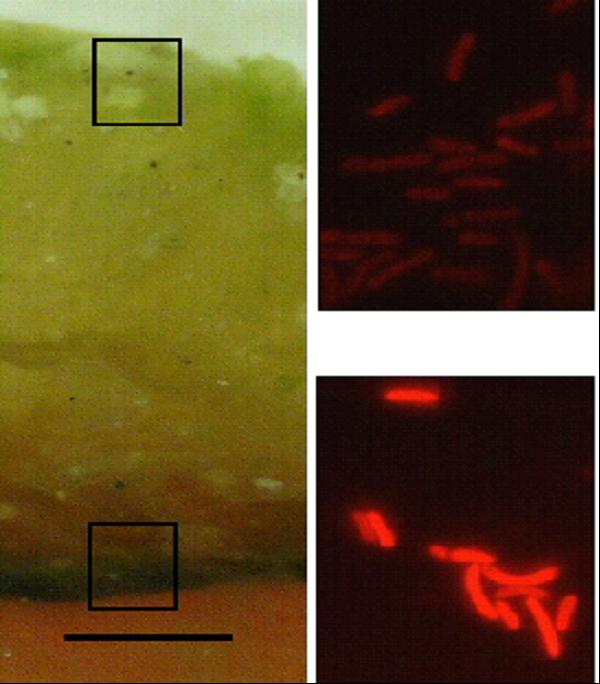
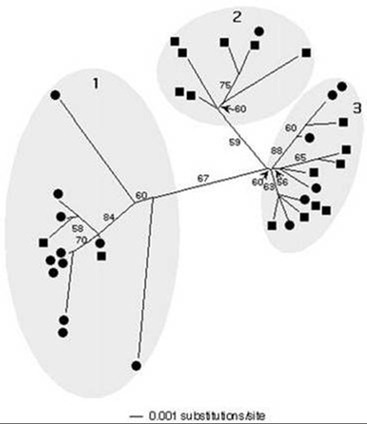
Figure 6. Left: PCR approach to amplification of 16s rRNA gene and adjacent internal transcribed spacer. Center: Vertical section of 68°C Mushroom Spring mat with autofluorescence microscopy images of surface and deeper layers. Right: Unrooted ITS phylogeny for cyanobacterial sequences of a single 16s rRNA type detected in the top layers (circles) or deeper layers (squares).
