Molecular analysis of community composition and structure
|
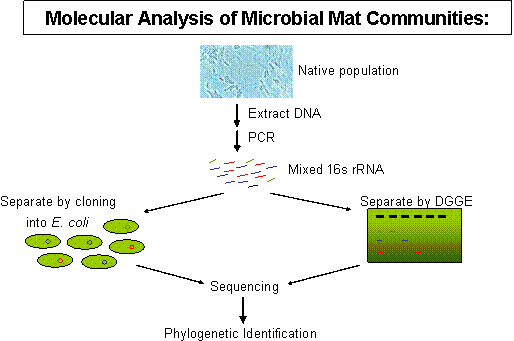
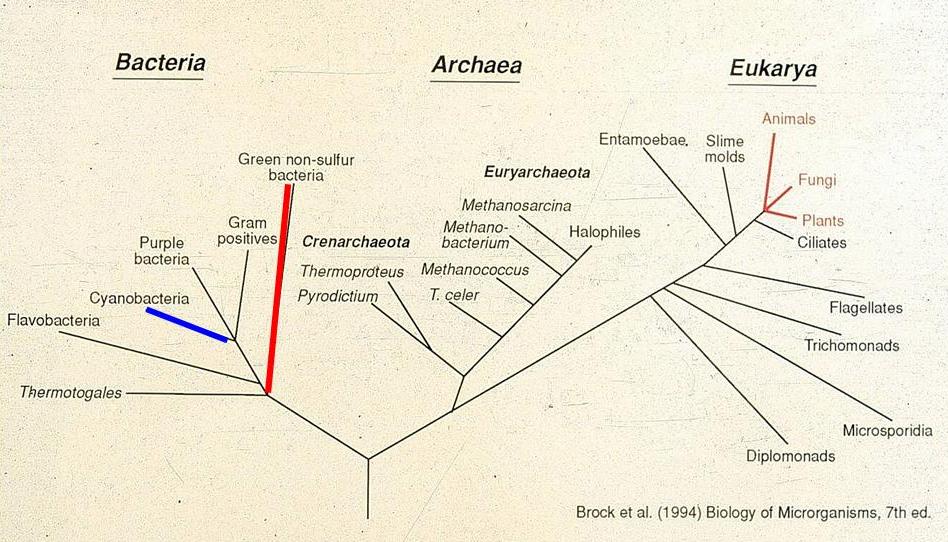
In the mid-1980s we began to develop and use methods for examining the genetic variation exhibited by the gene encoding 16S ribosomal RNA molecules, using the strategy depicted in Fig. 2. DNA is extracted from mat samples, then PCR amplified to produce a mixture of 16S rRNA sequences from community members. These are separated by cloning or gel methods calleddenaturing gradient gel electrophoresis (DGGE) to allow purification of individual 16S rRNA sequences. Purified 16S rRNAs are then sequenced and compared to a large and growing database on the evolution of life based on sequence variation in this moleculehttp://rdp.cme.msu.edu/index.jsp. As expected, our community contained 16S rRNA sequences related to cyanobacteria and green nonsulfur bacteria, as shown by the blue- and red- highlighted Kingdom-level lineages in the 3-domain tree. The three red-highlighted lineages in Domain Eukarya are the traditional Animal, Plant and Fungal Kingdoms. Since line length separating organisms in the tree are proportional to genetic difference between the organisms, it is easy to note that microorganisms (all other lines) contain much more genetic diversity as assayed by 16S rRNA sequence variation.
Figure 2. Approaches to 16s rRNA diversity analysis. |